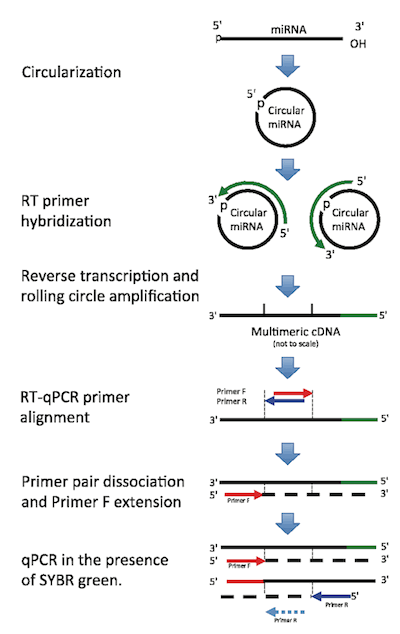
miR-ID® microRNA Detection Platform
miR-ID® is a novel platform for detecting miRNA using a circularization-based RT-qPCR method. miR-ID® is highly sensitive, uses single-dye detection, and can discriminate miRNA isoforms with single nucleotide differences at any position along the molecule. The technology works well with all sample sources, including total RNA, cell lysates, and tissue lysates.
miR-ID® provides better sensitivity, specificity, and versatility than competing methods. In addition, miR-ID® is the only PCR-based platform that is able to distinguish small RNAs with commonly occurring end modifications such as 5' triphosphates (e.g., secondary siRNAs in C. elegans), and 2'-OMe modifications (e.g., 3' ends of piRNAs in animals and all small RNAs in plants). Such end modifications are thought to play a role in both the stability and function of small RNAs, and there is a clear need to differentiate and detect modified small RNAs from their unmodified versions.
Comparison of miR-ID® and the leading competitor for small RNA detection
| miR-ID® | Leading Competitor Assays | |
|---|---|---|
| Sensitivity | ++++ | ++++ |
| Specificity | ++++ | ++ |
| Sequence coverage | ++++ | +++ |
| Distinguish and quantify RNA end modifications | Yes | No |
| Need for modified probes | No | Yes |
| Cost | $ | $$ |
| Discrimination between precursor and mature miRNAs | +++ | +++ |
| Works in cell lysates | Yes | Yes |
| Multiplex capability at RT step | Yes | Yes |
| Restrictions on primer position | None | Restricted to miRNA ends |
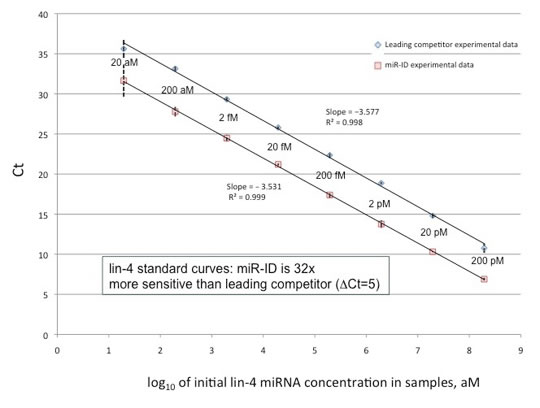
Sensitivity
The miR-ID® miRNA detection platform provides a higher signal-to-noise ratio than leading competitors, while using unmodified DNA primers, single-dye detection (SYBR green or EVA green), and no specialized probes. These features help keep the costs low and allow for a rapid development of assays for miRNAs of interest.
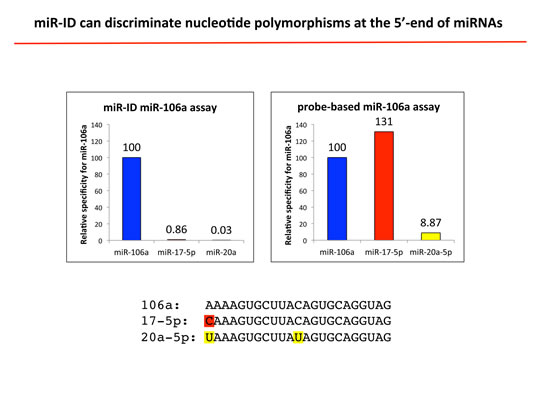
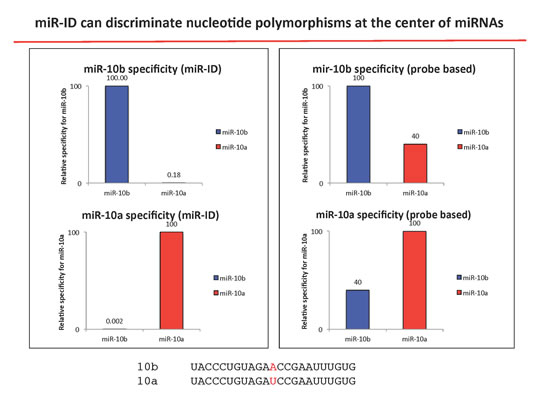
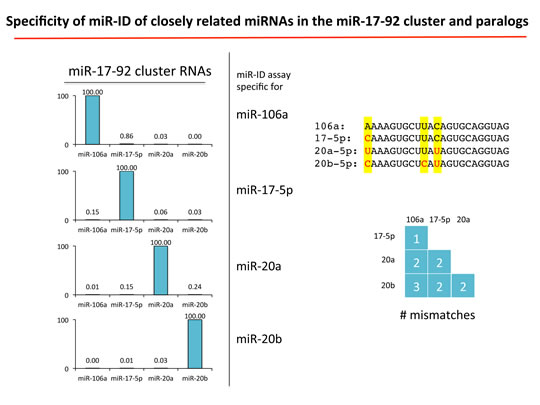
Specificity
miR-ID® provides unmatched discrimination between miRNA isoforms with single nucleotide differences. There is virtually no cross-talk between miRNAs from the same family, as well as superior discrimination between pre- and mature miRNAs.
Figure 3c: miR-ID® provides exceptional discrimination of closely related miRNAs of the miR-17-92 cluster. (click image to enlarge)
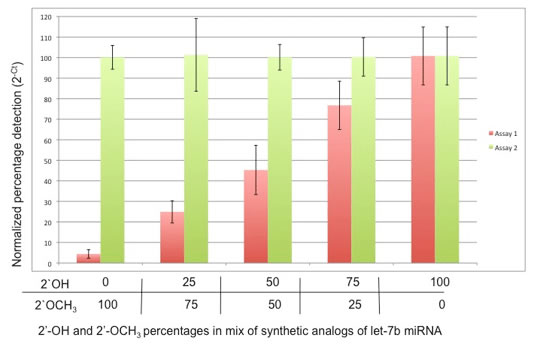
Quantification of End-Modified Small RNAs
miR-ID® is the first platform that can quantitatively discriminate between small RNAs with an identical sequence but differing terminal modifications. This is particularly useful in plant miRNAs, where discrimination between 2-O-methyl modified and unmodified miRNA is of considerable interest.
Convenience
miR-ID® can accommodate a variety of RNA sources without purification, including tissue or cell lysates. For the analysis of total RNA, no enrichment for small RNAs is required: miR-ID® works as well with total RNA as with enriched fractions.